
Multi-subject analyses can be accomplished by moving each subject to a standardized space and then performing voxel-wise or region-of-interest analyses across subjects. Optimal alignment methods can be dependent upon species, image modality and contrast, hardware, etc. Fastmap provides for manual and automated alignment to user-defined templates, while providing flexible methods to accomplish the process. A typical analysis process might include
- Brain extraction: This is an optional step. The method of brain extraction used in fastmap is quite simple and is not intended (and generally does not) give ideal results. If image contrast and extraction parameters are used to work in a single case, experience has shown this typically works in most similar use cases. In fastmap, the main purpose of brain extraction is to improve the “capture range” for automated adjustment based upon a cost function.
- FOV enlargement: This is a rarely used feature that most generally is used during selection of subjects from a multi-subject imaging cohort: e.g., multi-mouse imaging during PET or prenatal MRI in preclinical models. In such cases, one can enlarge the FOV, which is defined by the anatomical template, select the subject, and the shift the FOV to focus on the selected subject. When alignment is finished, use “save as” to save the result to a user-specified directory, and then use “time series —> reload time series” to reload the multi-subject time series and select the next subject.
- Orientation adjustment: It’s common to choose image planes that nearly match anatomical reference planes (coronal, sagittal, transverse), but these may differ from an anatomical template by 90 or 180 degree rotations, or even by a coordinate parity flip. These can and should be adjusted first manually before attempting automated alignment.
- Image flattening (or RF bias correction): Different MRI coils induce different amounts of regional signal variation (RF bias), which in turn can bias alignment. Fastmap divides the signal by a spatially smoothed version of itself to reduce long-range variations in intensity.
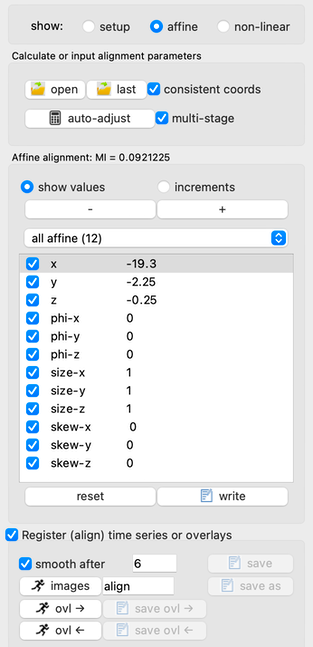
- Cost-function region: A typical approach in preclinical models is to rely upon the brain designation in anatomical space (either using an overlay as a mask or by setting non-brain voxels to zero in the template image) to remove all extravascular signal in the cost function as the source image is progressively moved into the anatomical space during automated alignment. When extra-cerebral signal has been removed from BOTH the template and the source volumes, it can be advantageous to use all voxels (checkbox at right). This will both increase the capture range in the automated alignment and also provide a degree of boundary-based optimization, which can be either good or bad based upon the quality of the brain extraction. When multi-stage alignment is selected (default), volumes will be progressively flattened at later alignment stages, and the source mask will be removed at late stages so that alignment relies only upon the template mask.
- Manual adjustment: In most cases, manual adjustment will not be needed unless the image quality is bad, the template and source are poorly matched, or the starting point of automated alignment is too far from the optimum. In this case, increments can be defined prior to manually adjusting values.
- Initialization: One can read parameters from a prior alignment within the same session (e.g, apply an anatomical alignment to PET) or use this feature to initialize alignment parameters prior to automated alignment. If the “consistent coords” box is checked, then a “reslice matrix” will be applied prior to the affine matrix to account for potentially different voxels with the same coordinates. E.g., one can align oblique EPI data or rotated PET data from an anatomical MRI scan within the same session.
- Auto-adjustment: The calculator button auto-adjusts alignment using Mutual Information as the cost function, subject to spatial restrictions enforced by the brain templates and spatial flattening as previously described. This step only determines parameters but does not alter any time-series data in program memory.
- Transform images: All images in all scans will be transformed to the template space by clicking the running man button. Images will be stored to respective directories using the prescribed name. Alternatively, “save as” will save the time series to user-defined directory; this feature, along with the menu function “Time Series —> reload time series”, is particularly useful for multi-subject imaging.
- Transform regions: Regions (overlays) can be transformed from either space to the other space using the affine alignment. These regions can then be stored to a user-defined directory.