Out:
Reading /home/ubuntu/mne_data/MNE-sample-data/MEG/sample/sample_audvis-ave.fif ...
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Left Auditory)
0 CTF compensation matrices available
nave = 55 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
Applying baseline correction (mode: mean)
# Author: Mainak Jas <mainak@neuro.hut.fi>
#
# License: BSD (3-clause)
import matplotlib.pyplot as plt
import mne
from mne.viz import tight_layout
from mne.datasets import sample
print(__doc__)
data_path = sample.data_path()
fname = data_path + '/MEG/sample/sample_audvis-ave.fif'
# Reading evoked data
condition = 'Left Auditory'
evoked = mne.read_evokeds(fname, condition=condition, baseline=(None, 0),
proj=True)
ch_names = evoked.info['ch_names']
picks = mne.pick_channels(ch_names=ch_names, include=["MEG 2332"])
# Create subplots
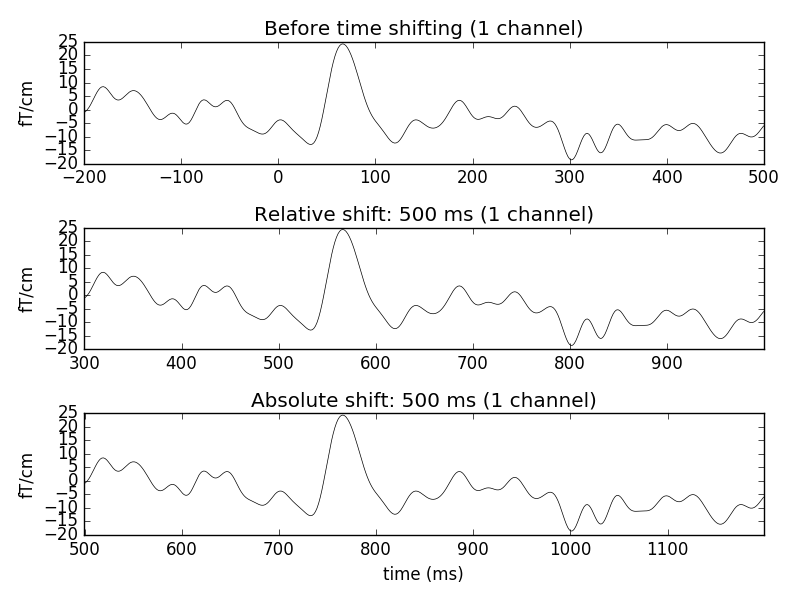
f, (ax1, ax2, ax3) = plt.subplots(3)
evoked.plot(exclude=[], picks=picks, axes=ax1,
titles=dict(grad='Before time shifting'))
# Apply relative time-shift of 500 ms
evoked.shift_time(0.5, relative=True)
evoked.plot(exclude=[], picks=picks, axes=ax2,
titles=dict(grad='Relative shift: 500 ms'))
# Apply absolute time-shift of 500 ms
evoked.shift_time(0.5, relative=False)
evoked.plot(exclude=[], picks=picks, axes=ax3,
titles=dict(grad='Absolute shift: 500 ms'))
tight_layout()
Total running time of the script: ( 0 minutes 0.823 seconds)