For a generic introduction to the computation of ERP and ERF see Epoching and averaging (ERP/ERF). Here we cover the specifics of EEG, namely:
- setting the reference
- using standard montages
mne.channels.Montage()- Evoked arithmetic (e.g. differences)
import mne
from mne.datasets import sample
Setup for reading the raw data
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
event_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
raw = mne.io.read_raw_fif(raw_fname, preload=True)
raw.set_eeg_reference() # set EEG average reference
Out:
An average reference projection was already added. The data has been left untouched.
Let’s restrict the data to the EEG channels
raw.pick_types(meg=False, eeg=True, eog=True)
By looking at the measurement info you will see that we have now 59 EEG channels and 1 EOG channel
print(raw.info)
Out:
<Info | 19 non-empty fields
bads : list | 0 items
buffer_size_sec : numpy.float64 | 13.3196808772
ch_names : list | EEG 001, EEG 002, EEG 003, EEG 004, EEG 005, EEG 006, ...
chs : list | 60 items (EEG: 59, EOG: 1)
comps : list | 0 items
custom_ref_applied : bool | False
dev_head_t : 'mne.transforms.Transform | 3 items
dig : list | 146 items
events : list | 0 items
file_id : dict | 4 items
highpass : float | 0.10000000149 Hz
hpi_meas : list | 1 items
hpi_results : list | 1 items
lowpass : float | 40.0 Hz
meas_date : numpy.ndarray | 2002-12-03 19:01:10
meas_id : dict | 4 items
nchan : int | 60
projs : list | PCA-v1: off, PCA-v2: off, PCA-v3: off, ...
sfreq : float | 150.153747559 Hz
acq_pars : NoneType
acq_stim : NoneType
ctf_head_t : NoneType
description : NoneType
dev_ctf_t : NoneType
experimenter : NoneType
hpi_subsystem : NoneType
kit_system_id : NoneType
line_freq : NoneType
proj_id : NoneType
proj_name : NoneType
subject_info : NoneType
xplotter_layout : NoneType
>
In practice it’s quite common to have some EEG channels that are actually
EOG channels. To change a channel type you can use the
mne.io.Raw.set_channel_types() method. For example
to treat an EOG channel as EEG you can change its type using
raw.set_channel_types(mapping={'EOG 061': 'eeg'})
print(raw.info)
Out:
<Info | 19 non-empty fields
bads : list | 0 items
buffer_size_sec : numpy.float64 | 13.3196808772
ch_names : list | EEG 001, EEG 002, EEG 003, EEG 004, EEG 005, EEG 006, ...
chs : list | 60 items (EEG: 60)
comps : list | 0 items
custom_ref_applied : bool | False
dev_head_t : 'mne.transforms.Transform | 3 items
dig : list | 146 items
events : list | 0 items
file_id : dict | 4 items
highpass : float | 0.10000000149 Hz
hpi_meas : list | 1 items
hpi_results : list | 1 items
lowpass : float | 40.0 Hz
meas_date : numpy.ndarray | 2002-12-03 19:01:10
meas_id : dict | 4 items
nchan : int | 60
projs : list | PCA-v1: off, PCA-v2: off, PCA-v3: off, ...
sfreq : float | 150.153747559 Hz
acq_pars : NoneType
acq_stim : NoneType
ctf_head_t : NoneType
description : NoneType
dev_ctf_t : NoneType
experimenter : NoneType
hpi_subsystem : NoneType
kit_system_id : NoneType
line_freq : NoneType
proj_id : NoneType
proj_name : NoneType
subject_info : NoneType
xplotter_layout : NoneType
>
And to change the nameo of the EOG channel
raw.rename_channels(mapping={'EOG 061': 'EOG'})
Let’s reset the EOG channel back to EOG type.
raw.set_channel_types(mapping={'EOG': 'eog'})
The EEG channels in the sample dataset already have locations. These locations are available in the ‘loc’ of each channel description. For the first channel we get
print(raw.info['chs'][0]['loc'])
Out:
[-0.03737009 0.10568011 0.07333875 0.00235201 0.11096951 -0.03500458
0. 1. 0. 0. 0. 1. ]
And it’s actually possible to plot the channel locations using
the mne.io.Raw.plot_sensors() method
raw.plot_sensors()
raw.plot_sensors('3d') # in 3D
In the case where your data don’t have locations you can set them
using a mne.channels.Montage(). MNE comes with a set of default
montages. To read one of them do:
montage = mne.channels.read_montage('standard_1020')
print(montage)
Out:
<Montage | standard_1020 - 97 channels: LPA, RPA, Nz ...>
To apply a montage on your data use the set_montage method.
function. Here don’t actually call this function as our demo dataset
already contains good EEG channel locations.
Next we’ll explore the definition of the reference.
Let’s first remove the reference from our Raw object.
This explicitly prevents MNE from adding a default EEG average reference required for source localization.
raw_no_ref, _ = mne.set_eeg_reference(raw, [])
We next define Epochs and compute an ERP for the left auditory condition.
reject = dict(eeg=180e-6, eog=150e-6)
event_id, tmin, tmax = {'left/auditory': 1}, -0.2, 0.5
events = mne.read_events(event_fname)
epochs_params = dict(events=events, event_id=event_id, tmin=tmin, tmax=tmax,
reject=reject)
evoked_no_ref = mne.Epochs(raw_no_ref, **epochs_params).average()
del raw_no_ref # save memory
title = 'EEG Original reference'
evoked_no_ref.plot(titles=dict(eeg=title))
evoked_no_ref.plot_topomap(times=[0.1], size=3., title=title)
Average reference: This is normally added by default, but can also be added explicitly.
raw_car, _ = mne.set_eeg_reference(raw)
evoked_car = mne.Epochs(raw_car, **epochs_params).average()
del raw_car # save memory
title = 'EEG Average reference'
evoked_car.plot(titles=dict(eeg=title))
evoked_car.plot_topomap(times=[0.1], size=3., title=title)
Out:
An average reference projection was already added. The data has been left untouched.
Custom reference: Use the mean of channels EEG 001 and EEG 002 as a reference
raw_custom, _ = mne.set_eeg_reference(raw, ['EEG 001', 'EEG 002'])
evoked_custom = mne.Epochs(raw_custom, **epochs_params).average()
del raw_custom # save memory
title = 'EEG Custom reference'
evoked_custom.plot(titles=dict(eeg=title))
evoked_custom.plot_topomap(times=[0.1], size=3., title=title)
Trial subsets from Epochs can be selected using ‘tags’ separated by ‘/’. Evoked objects support basic arithmetic. First, we create an Epochs object containing 4 conditions.
event_id = {'left/auditory': 1, 'right/auditory': 2,
'left/visual': 3, 'right/visual': 4}
epochs_params = dict(events=events, event_id=event_id, tmin=tmin, tmax=tmax,
reject=reject)
epochs = mne.Epochs(raw, **epochs_params)
print(epochs)
Out:
<Epochs | n_events : 288 (good & bad), tmin : -0.199795213158 (s), tmax : 0.499488032896 (s), baseline : (None, 0), ~3.1 MB, data not loaded,
'left/auditory': 72, 'left/visual': 73, 'right/auditory': 73, 'right/visual': 70>
Next, we create averages of stimulation-left vs stimulation-right trials. We can use basic arithmetic to, for example, construct and plot difference ERPs.
left, right = epochs["left"].average(), epochs["right"].average()
# create and plot difference ERP
mne.combine_evoked([left, -right], weights='equal').plot_joint()
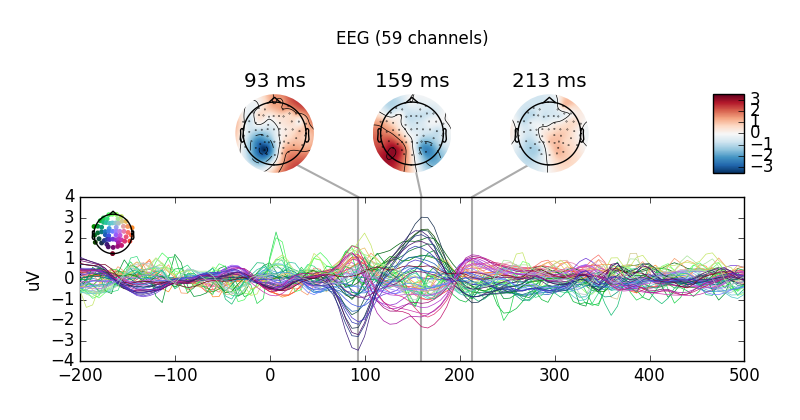
This is an equal-weighting difference. If you have imbalanced trial numbers,
you could also consider either equalizing the number of events per
condition (using
epochs.equalize_event_counts).
As an example, first, we create individual ERPs for each condition.
aud_l = epochs["auditory", "left"].average()
aud_r = epochs["auditory", "right"].average()
vis_l = epochs["visual", "left"].average()
vis_r = epochs["visual", "right"].average()
all_evokeds = [aud_l, aud_r, vis_l, vis_r]
print(all_evokeds)
Out:
[<Evoked | comment : 'auditory+left', kind : average, time : [-0.199795, 0.499488], n_epochs : 185, n_channels x n_times : 59 x 106, ~3.1 MB>, <Evoked | comment : 'auditory+right', kind : average, time : [-0.199795, 0.499488], n_epochs : 174, n_channels x n_times : 59 x 106, ~3.1 MB>, <Evoked | comment : 'visual+left', kind : average, time : [-0.199795, 0.499488], n_epochs : 179, n_channels x n_times : 59 x 106, ~3.1 MB>, <Evoked | comment : 'visual+right', kind : average, time : [-0.199795, 0.499488], n_epochs : 185, n_channels x n_times : 59 x 106, ~3.1 MB>]
This can be simplified with a Python list comprehension:
all_evokeds = [epochs[cond].average() for cond in sorted(event_id.keys())]
print(all_evokeds)
# Then, we construct and plot an unweighted average of left vs. right trials
# this way, too:
mne.combine_evoked(all_evokeds,
weights=(0.25, -0.25, 0.25, -0.25)).plot_joint()
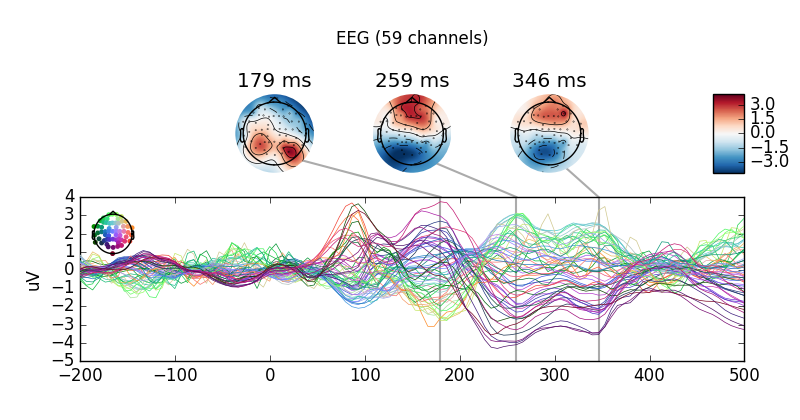
Out:
[<Evoked | comment : 'left/auditory', kind : average, time : [-0.199795, 0.499488], n_epochs : 56, n_channels x n_times : 59 x 106, ~3.1 MB>, <Evoked | comment : 'left/visual', kind : average, time : [-0.199795, 0.499488], n_epochs : 67, n_channels x n_times : 59 x 106, ~3.1 MB>, <Evoked | comment : 'right/auditory', kind : average, time : [-0.199795, 0.499488], n_epochs : 62, n_channels x n_times : 59 x 106, ~3.1 MB>, <Evoked | comment : 'right/visual', kind : average, time : [-0.199795, 0.499488], n_epochs : 56, n_channels x n_times : 59 x 106, ~3.1 MB>]
Often, it makes sense to store Evoked objects in a dictionary or a list - either different conditions, or different subjects.
# If they are stored in a list, they can be easily averaged, for example,
# for a grand average across subjects (or conditions).
grand_average = mne.grand_average(all_evokeds)
mne.write_evokeds('/tmp/tmp-ave.fif', all_evokeds)
# If Evokeds objects are stored in a dictionary, they can be retrieved by name.
all_evokeds = dict((cond, epochs[cond].average()) for cond in event_id)
print(all_evokeds['left/auditory'])
# Besides for explicit access, this can be used for example to set titles.
for cond in all_evokeds:
all_evokeds[cond].plot_joint(title=cond)
Out:
<Evoked | comment : 'left/auditory', kind : average, time : [-0.199795, 0.499488], n_epochs : 56, n_channels x n_times : 59 x 106, ~3.1 MB>
Total running time of the script: ( 0 minutes 14.115 seconds)