jip displays voxels
The display orientation of a volume depends only upon the voxel indices and program settings, and not upon the coordinate system attached to the voxels. This is unlike some other programs that assume a standard coordinate convention for humans, which can lead to rotations in the display that depend both upon both voxels indices and coordinates.
The position of 4-dimension voxel (i j k t) within a volume of dimension (Nx Ny Nz Nt) is given by t*Nx*Ny*Nz + k*Nx*Ny + j*Ny + i, which is to say that the x dimension moves fastest, the y dimension 2nd fastest, and so on. This is the standard convention for voxel ordering.
In mosaic display mode, voxels in the i-j plane are shown, with the k index start at 0 in the upper left panel and increasing in panels to the right and down. Within each panel, the (i j) = (0 0) voxel is shown at the bottom left unless the "-y" option is used on startup in that case, the origin is the top left (e.g., the y dimension is swapped up-down). The first convention is useful for displaying files that use the X11 convention, where the image origin is the top left pixel, or for displaying data using the DICOM convention, which is rotated by 180 degrees with respect to the NIFTI convention. The templates provided with the jip package are designed to display correctly without the "-y" option, meaning that a rotation often will be required during alignment. This is somewhat inconvenient, but the up-down orientation was selected for compatibility with other display programs and, as much as possible, with the intention of NIFTI. Use “y” interactively to swap up-down conventions.
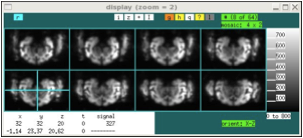
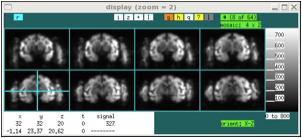
xd data-from-scanner.nii xd data-from-scanner.nii -y
Note that mosaic mode can switch between display of three orthogonal orientations (for aligned data, these are coronal, sagital, and axial) by selecting the “orient” field below the image window. Also, the number of panels can be restricted (top right, “#”), or the panels can be selected with mouse clicks in order to display non-contiguous slices. When displaying a restricted slice set, the mouse scroll wheel or the “< >” arrow keys can be used to move through the slice set.
In tri-planar display mode, there are two different "standard" display orientation that can be used after loading data. These 2 modes are used to accommodate both rodent and human/primate data, which generally use different conventions as a result of the different orientations of the brains in the scanner, as well as the fact that most rodent histology is displayed in the coronal orientation. Most rodent MRI data is collected in the coronal view, but most NIFTI display packages assume that the i-j plane is an axial view of the human brain. In short, display rodent data in the default mode with "Y vs Z" as the text field above the top right part of the display. For human or non-human primate data, switch this orientation to "Z vs Y". The default tri-planar orientation is designed for rodents. To change the default on program entry, use the "-Y" option. If this is the common usage, set this as "alias xd ~/jip-current/bin/display -Y".
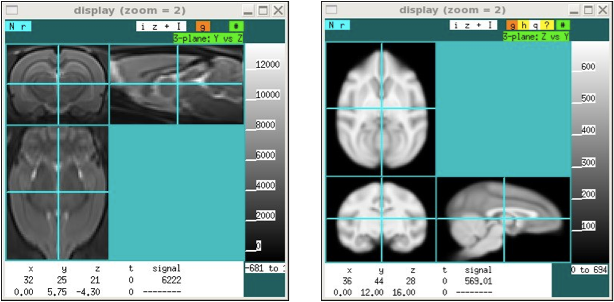
xd rat.nii -3 xd monkey.nii -3 -Y