The brain extraction method is designed primarily for preclinical models, specifically to help facilitate spatial normalization (alignment) to brain templates. It is not intended to provide a particularly accurate representation; rather, it’s meant to improve the capture range of the first stage of automated alignment. After the first stage, one can rely upon the template delineation to exclude extra cerebral tissue. This brain extraction method can also be accessed directly from the alignment preprocessing panel; in multi-stage alignment, the subject-specific brain region is eliminated in later stages in favor of reliance on the template brain region only.
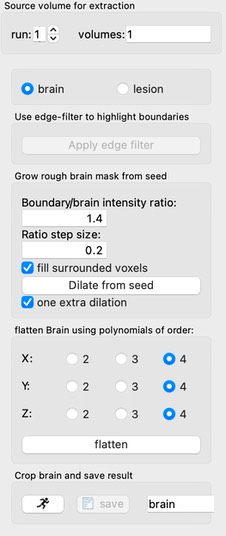
This method first employs an edge filter, followed by restricted dilation from a seed. The seed is defined as the crosshair position, and dilation proceeds subject to a boundary ratio defined relative to the median of the signal within the dilating brain mask. The boundary ratio is slowly escalated. Following a rapid growth phase, the growth rate decreases, and the process is terminated when it again increases (often indicating escape).
This process is subject to errors due to signal inhomogeneity, poor quality, or restricted brain slices. Note that this method actually works best with low contrast (e.g., short echo times), since it relies upon more contrast at the brain edge than within the brain. If one desires more accurate segmentation of brain for individual subjects, an alternative is template-based alignment, followed by an inverse transform of the template brain region.
Note this this method is relatively fast in lower species but becomes slow at high resolution with large field of view.
The same method can be used for lesion segmentation with negative contrast (e.g., hematoma). In such a scenario, one can first extract the brain and then then flatten the signal (remove spatial variation) using a polynomial fit.